Category: Parkinson's Disease: Genetics
Objective: To assess genome-wide genetic risk for Parkinson’s disease (PD) within and across genetically determined European ancestries (European, Ashkenazi Jewish, and Finnish) from the Global Parkinson’s Genetic Program (GP2, http://gp2.org/), previously published PD GWAS (IPDGC), the Fox Insight Genetics Study (FIGS), and national biobanks FinnGen and the UK Biobank (UKB).
Background: Genome-wide association studies (GWAS) in PD have significantly expanded our biological understanding of the disease, with previous studies nominating up to 90 genetic risk variants.1 GWAS of increasing sample size and quality of genotyping and sequencing will be important for identifying additional variants that can provide further insight into PD genetics. Additionally, previous research has suggested that allele frequencies and linkage disequilibrium (LD) patterns can differ among European sub-populations at disease risk loci.2 Employing advanced genetic ancestry estimates for European populations allows us to investigate risk at a more detailed level, aiding to expand our knowledge of the architecture of PD risk.
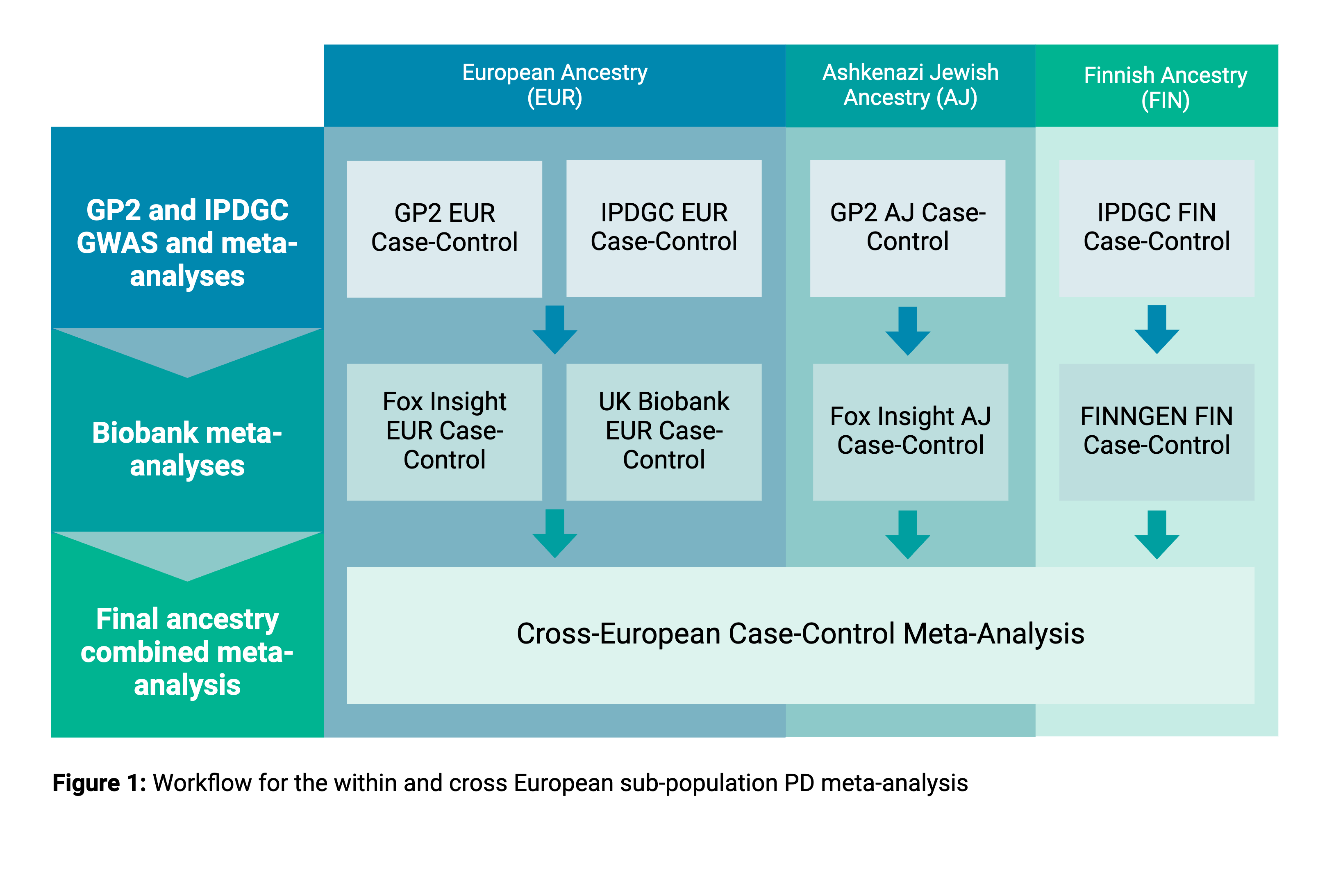
Method: We performed a series of GWAS using genotyped imputed data from GP2, IPDGC, Fox Insight, and UKB within and across three ancestry groups: European (EUR), Ashkenazi Jewish (AJ), and Finnish (FIN). Genetically-defined ancestry groups for GP2, IPDGC, and Fox Insight were defined by the GP2 ancestry and QC pipeline. EUR ancestry for UKB was defined by ancestry clustering performed by UKB. FinnGen PD summary statistics were downloaded from the Risteys portal. Meta-analyses were performed in stages, splitting by ancestry and biobank data sources (Figure 1). The last stage combined all European ancestry groups in a final pan-European meta-analysis.
Results: In our preliminary analysis, we identified several potential novel loci (P<5e-8) both within and across ancestries. When comparing results across ancestry groups, known risk loci often had different lead variants and frequencies. Known risk loci also displayed differing LD patterns, such as for LRRK2 in the AJ ancestry.
Conclusion: Our findings highlight the importance of exploring and comparing PD genetic risk in more granular populations, as well as the continuing need for larger GWAS with improved genotyping and imputation to nominate additional risk factors and broaden our knowledge of disease etiology.
Figure 1
References: 1. Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019).
2. Rivas, M. A. et al. Insights into the genetic epidemiology of Crohn’s and rare diseases in the Ashkenazi Jewish population. PLoS Genet. 14, e1007329 (2018).
To cite this abstract in AMA style:
G. Lobal_parkinson'S_genetics_program. Assessing Genome-wide Genetic Risk for Parkinson’s Disease in European Sub-Populations Across the Global Parkinson’s Genetics Program and National Biobanks [abstract]. Mov Disord. 2024; 39 (suppl 1). https://www.mdsabstracts.org/abstract/assessing-genome-wide-genetic-risk-for-parkinsons-disease-in-european-sub-populations-across-the-global-parkinsons-genetics-program-and-national-biobanks/. Accessed July 18, 2025.« Back to 2024 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/assessing-genome-wide-genetic-risk-for-parkinsons-disease-in-european-sub-populations-across-the-global-parkinsons-genetics-program-and-national-biobanks/