Category: Parkinson's Disease: Genetics
Objective: To identify novel monogenic causes of Parkinson’s disease (PD) by performing short and long-read sequencing for up to 10,000 PD patients and families with an unsolved but apparently monogenic cause of disease. Further, to improve current genotype-phenotype correlates and better understand variable penetrance and expressivity of pathogenic variants in known PD genes.
Background: The Global Parkinson’s Genetics Program (GP2, https://gp2.org/) is an international collaborative effort that aims to improve our understanding of the role genetics plays in PD [1]. The two main arms of GP2 are the “Monogenic Hub” and the “Complex Disease Hub”.
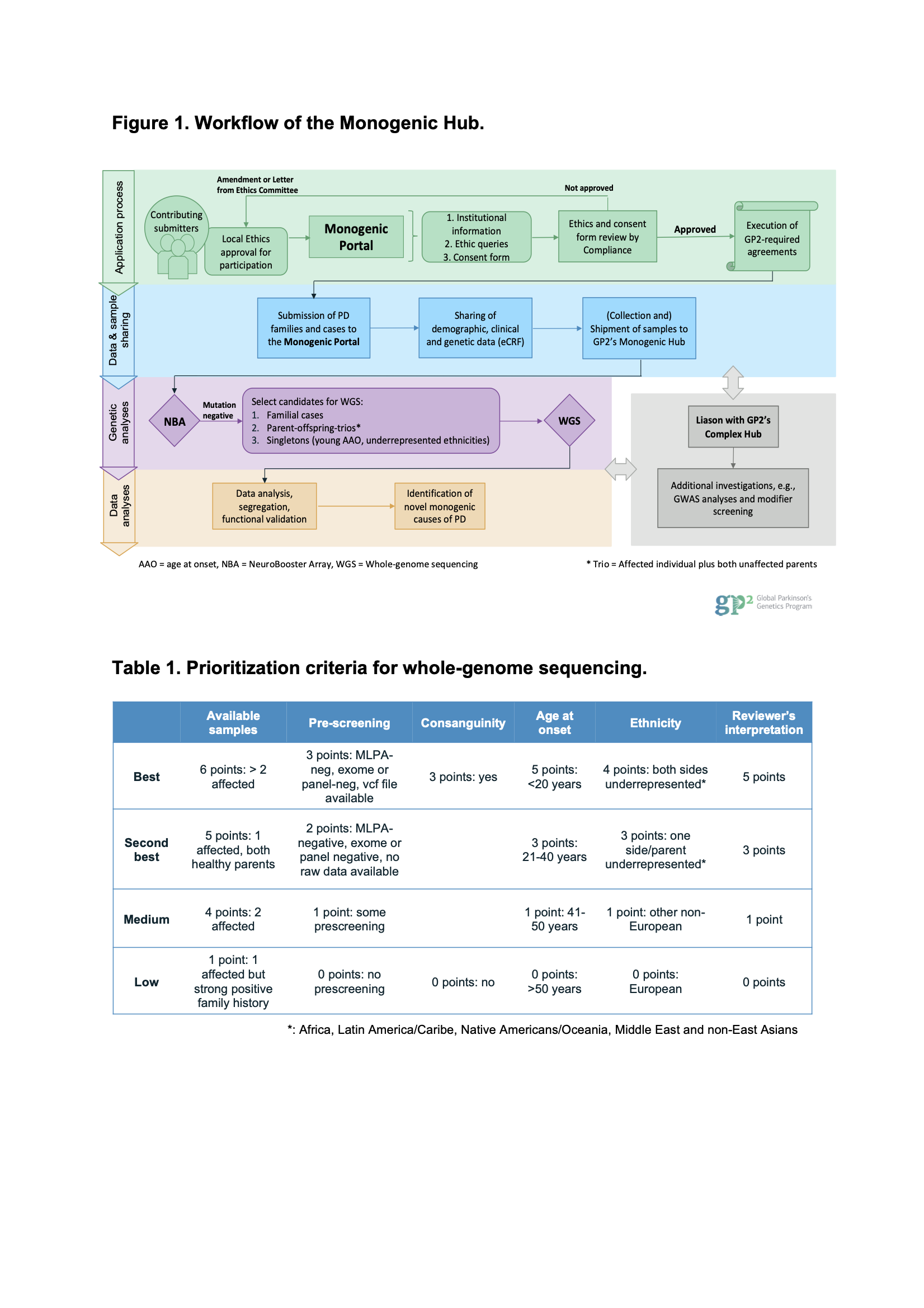
Method: [Figure1]: By reaching out to PD clinicians and researchers worldwide, we collect PD patients from multiplex families or as singleton cases, in whom a monogenic cause is suspected. We developed the GP2 Monogenic Portal, an easy-to-use online application that includes an electronic case report form (eCRF) to collect demographic, clinical, and pre-existing genetic data to help critically interpret genomic findings. All submitted cases undergo genotyping with the NeuroBooster Array. The array includes 1.9 million markers from the Illumina Global Diversity Array and over 95,000 neurological disease-oriented and population-specific variants (https://github.com/GP2code/Neuro_Booster_Array). It serves as a low-cost screening tool for variants in known PD genes. Mutation-negative cases are prioritized for whole-genome sequencing (WGS) based on different criteria [Table1].
Results: To date, the Monogenic Hub team has contacted more than 200 potential contributors from over 60 different countries, of whom 54 completed registration and 22 contracting. Genotyping and WGS have already been performed for >300 samples and are ongoing for another ~250. To date, genotyping of included patients with the NeuroBooster did not identify any carriers of known pathogenic PD gene variants, likely related to the thorough prescreening for most of the samples. WGS revealed the first novel candidate genes that are currently undergoing further investigation.
Conclusion: Our progress to date reflects a strong level of enthusiasm for the project. Together with the extensive infrastructure and global research of this project, we anticipate this study will identify novel PD genes and make a substantial contribution to the field of monogenic PD research.
References: [1] The Global Parkinson’s Genetics Program. GP2: The Global Parkinson’s Genetics Program. Mov Disord. First published: 29 January 2021. https://doi.org/10.1002/mds.28494.
To cite this abstract in AMA style:
L. Lange, ZH. Fang, E. Valente, SY. Lim, A. Tan, A. Illarionova, M. Avenali, I. Keller Sarmiento, H. Madoev, K. Roopnarain, C. Galandra, M. Ellis, K. Kumar, P. Heutink, N. Mencacci, K. Lohmann, C. Klein. Monogenic Hub of the Global Parkinson’s Genetics Program (GP2): In search of new PD genes [abstract]. Mov Disord. 2022; 37 (suppl 2). https://www.mdsabstracts.org/abstract/monogenic-hub-of-the-global-parkinsons-genetics-program-gp2-in-search-of-new-pd-genes/. Accessed June 30, 2025.« Back to 2022 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/monogenic-hub-of-the-global-parkinsons-genetics-program-gp2-in-search-of-new-pd-genes/