Category: Neuroimaging (Non-PD)
Objective: Establish a fast and reliable automatic tool for cerebellar subsegmentation.
Background: Quantifying the volume of the cerebellum and its lobes is of profound interest in various neurodegenerative and acquired diseases. Especially for the most common spinocerebellar ataxias (SCA), for which the first gene therapy interventional studies will start in the near future, there is an urgent need for quantitative, sensitive imaging markers at presymptomatic stages for stratification and treatment assessment.
Method: For training, validation, and testing of our deep learning method for cerebellar subsegmentation, CerebNet, T1-weighted images from 30 subjects were manually annotated into cerebellar lobules and vermal subsegments as well as cerebellar white matter. Our approach uses a UNet-based 2.5D segmentation network. We evaluated the performance with established metrics (Dice, Hausdorff Distance, 95th percentile of Hausdorff distance as well as volume similarity) and compared the performance with other state-of-the-art methods. Moreover, we applied the cerebellar subsegmentation in several ataxia cohorts (SCA1, 2, 3, 6 and MSA-C) as well as a large group of healthy controls along the entire lifespan (N=292).
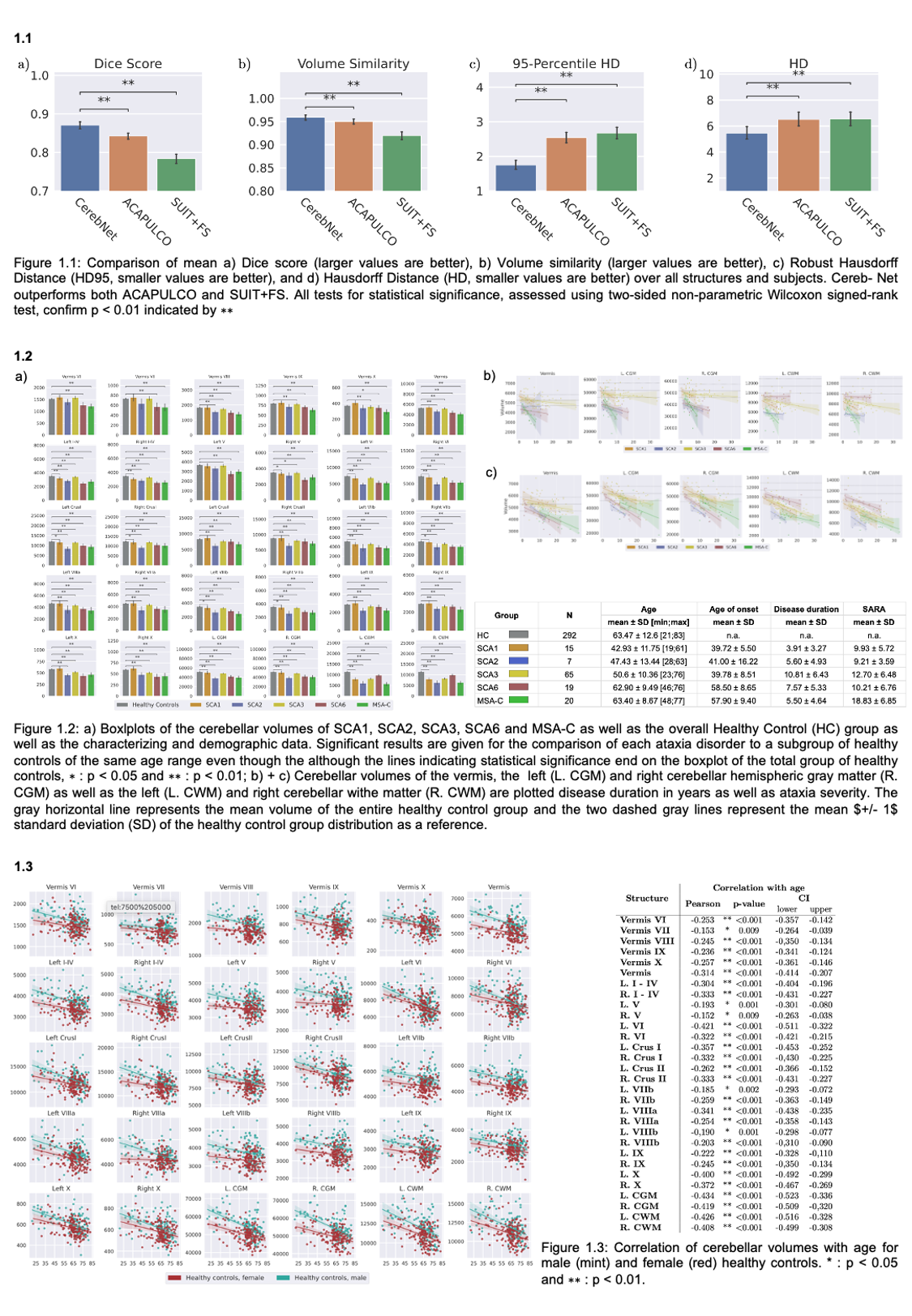
Results: CerebNet demonstrates a high accuracy (DICE = 0.87 and Hausdorf-95=1.75mm on average over all structures) outperforming state-of-the art approaches (Fig 1.1). Furthermore, it shows high test-retest reliability as well as high sensitivity to disease effects in SCA1, SCA2, SCA3, SCA6 and MSA-C (Fig. 1.2 and 1.3).
Conclusion: With CerebNet, we provide a fully automated and efficient deep learning method with extensive validation. CerebNet does not require any preprocessing, such as spatial normalization or bias field correction. It demonstrates a high segmentation accuracy outperforming other state-of-the-art approaches and has shown to be sensitive to capture disease and age-related changes in SCA1, SCA2, SCA3, SCA6 and MSA-C as well as healthy controls. CerebNet is compatible with FreeSurfer and FastSurfer and capable of rapidly processing a T1-weighted MRI within 12 seconds on a single GPU. This enables detailed studies of the cerebellum substructures and efficient evaluation even in very large cohorts. CerebNet will be available as an open-source project with a docker container at github.
To cite this abstract in AMA style:
J. Faber, D. Kuegler, E. Bahrami, L. Heinz, D. Timmann, T. Ernst, K. Deike-Hofmann, B. Vande Warrenburg, G. Oz, J. van Gaalen, J. Joers, K. Reetz, T. Klockgether, M. Reuter. CerebNet: Deep learning cerebellar subsegmentation for fast and reliable atrophy quantification [abstract]. Mov Disord. 2022; 37 (suppl 2). https://www.mdsabstracts.org/abstract/cerebnet-deep-learning-cerebellar-subsegmentation-for-fast-and-reliable-atrophy-quantification/. Accessed July 18, 2025.« Back to 2022 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/cerebnet-deep-learning-cerebellar-subsegmentation-for-fast-and-reliable-atrophy-quantification/