Category: Parkinson's Disease: Neuroimaging
Objective: Using MRI data, the aim of this longitudinal study was to map the progression of brain atrophy over four years and relate it to brain structural and functional connectivity and gene ontology enrichment analyses.
Background: Atrophy in multiple brain regions has been reported in the early stages of Parkinson’s Disease (PD) [1], but less is known about its progression. With few exceptions [2], no study has attempted to relate the pattern of atrophy progression to intrinsic brain properties such as anatomical connectivity and gene expression.
Method: Structural MRI and clinical data were obtained from the PPMI database [3] for 74 de novo PD patients (50 Men: 24 Women) with T1-weighted MRI at baseline, one, two and four years and 157 healthy control (HC; 115 Men: 42 Women) at baseline only. Deformation-based morphometry measure was used to quantify brain tissue atrophy [4] and W-score maps were computed to regress out the expected effects of age and sex using HC data [5]. We next investigated if the regional atrophy progression was correlated with the atrophy progression of its structurally and functionally connected neighborhood using Cammoun atlas [6] at four spatial resolutions (68, 114, 219 and 448) and networks from healthy adults [7]. We also investigated if the atrophy progression was associated with specific biological processes using a GO enrichment analysis and the Gorilla [8] and PANTHER [9] platforms.
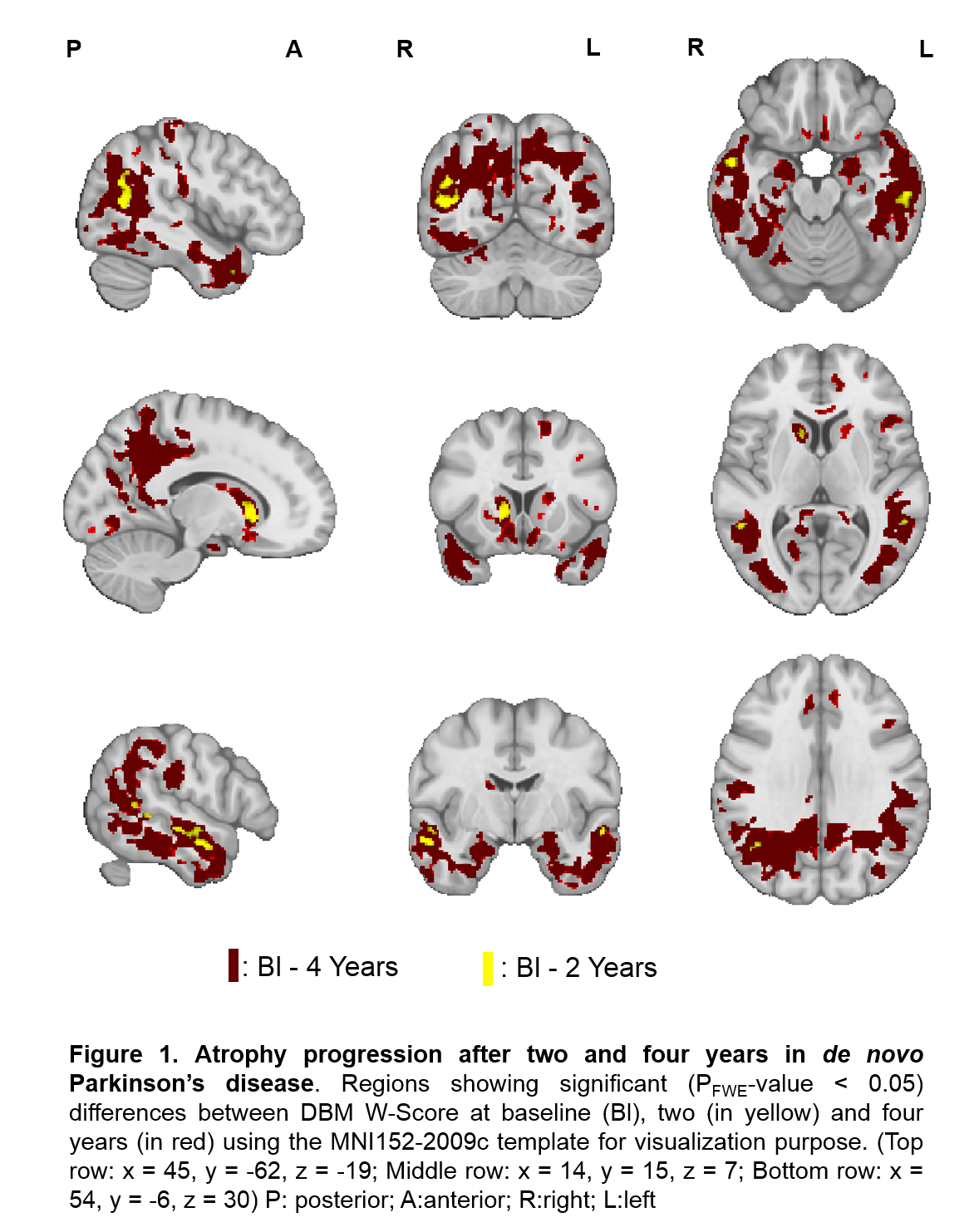
Results: We found that atrophy significantly progressed after two and four years in the caudate, nucleus accumbens, hippocampus, and different regions of the temporal, parietal, occipital and posterior cingulate cortex (Fig.1).
[figure1]
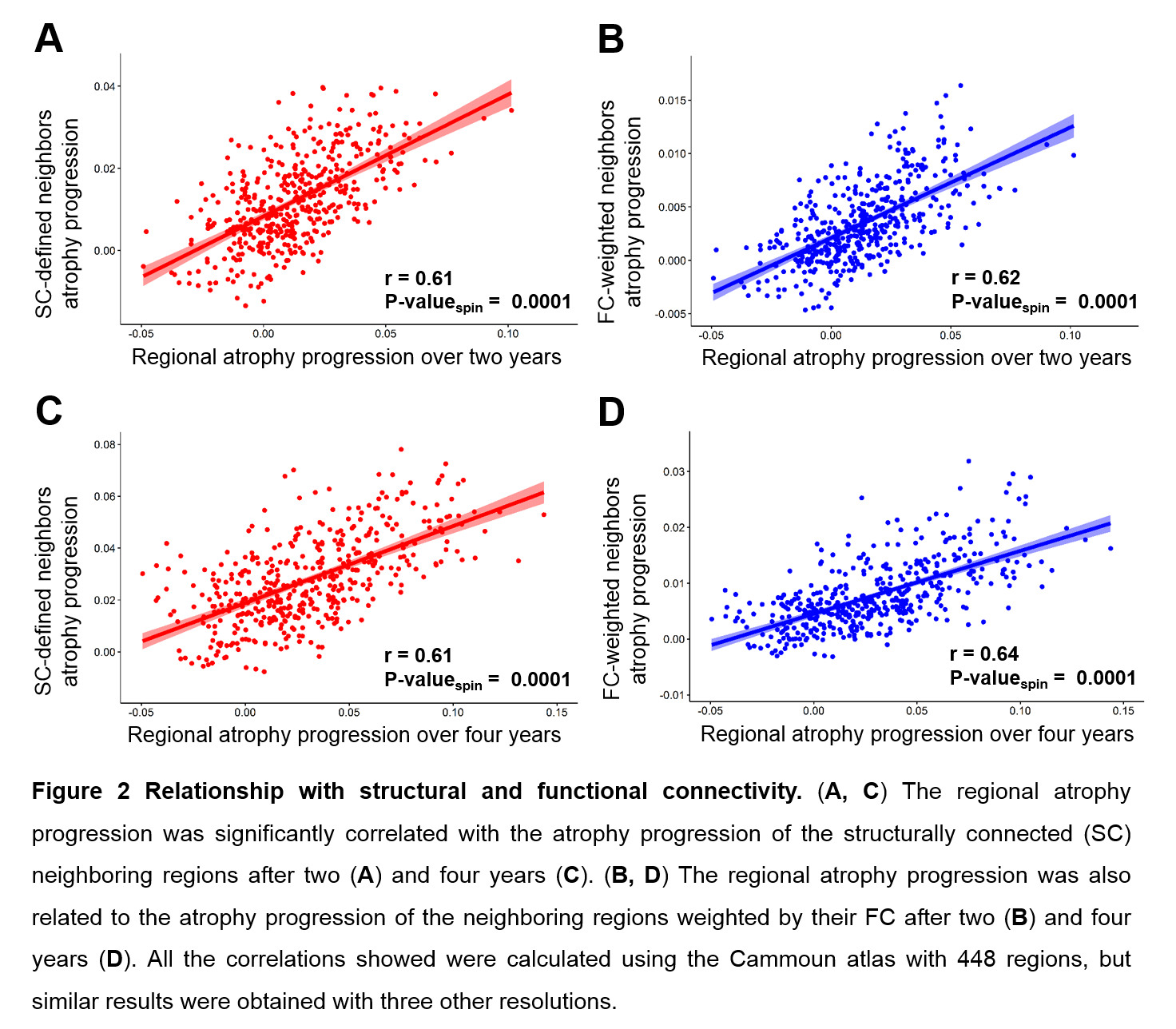
This progression was shaped by both structural and functional brain connectivity (Fig.2). Similar results were obtained at all the spatial resolutions.
[figure2]
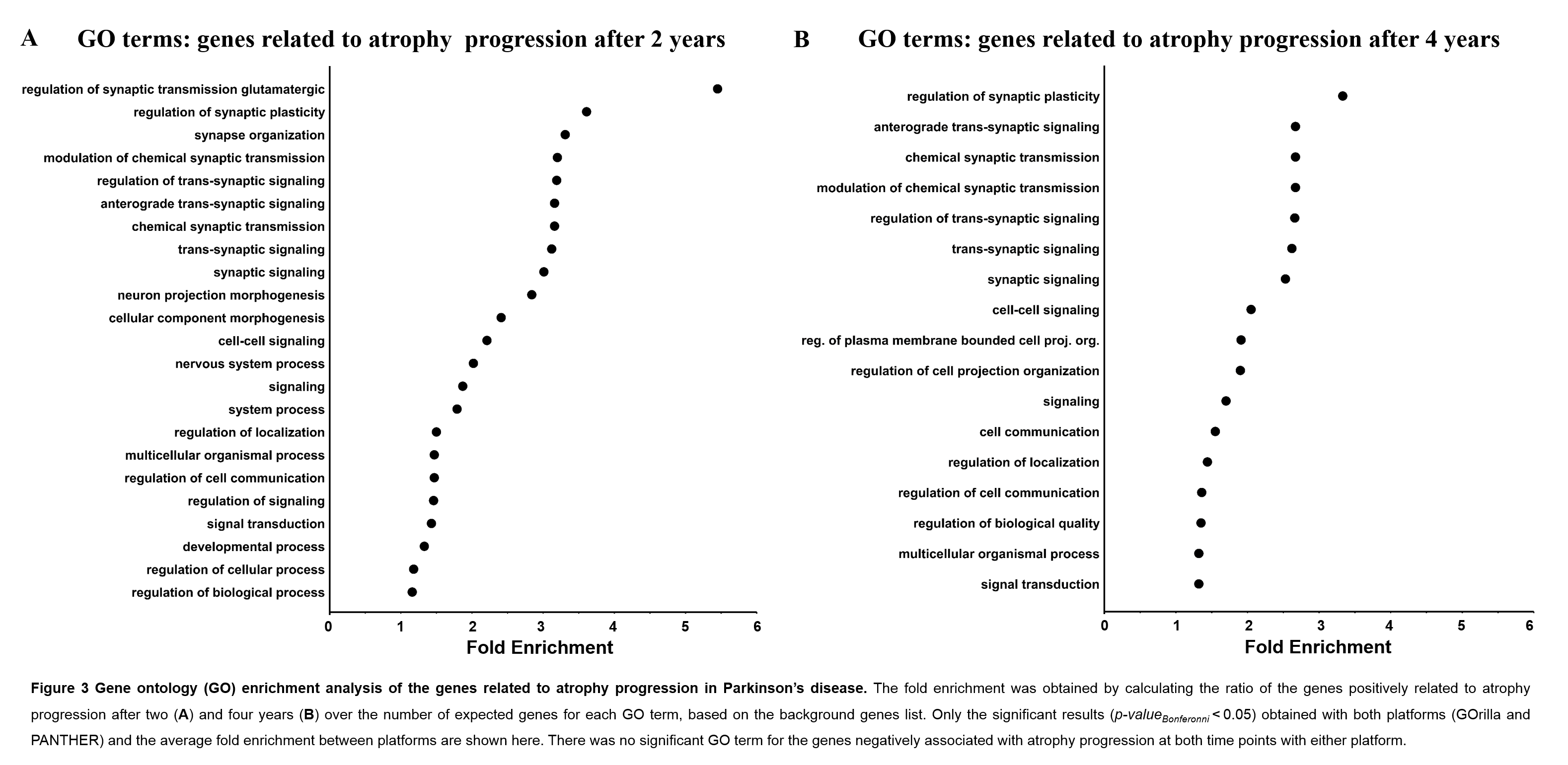
Also, the results from each platform consistently showed that the progression of atrophy after two and four years was more pronounced in regions with a higher expression of genes related to synaptic activity and signaling (Fig.3).
[figure3]
Conclusion: In sum, we demonstrate that the progression of atrophy in PD is in line with the prion-like propagation hypothesis of alpha-synuclein and provide evidence that synapses may be especially vulnerable to synucleinopathy. This abstract has been accepted for a presentation during the 2021 OHBM Annual Meeting.
References: [1]: Zeighami Y, Ulla M, Iturria-Medina Y, et al. Network structure of brain atrophy in de novo Parkinson’s disease. eLife. 2015;4(e08440):1-20. doi:10.7554/eLife.08440.001 [2]: Yau Y, Zeighami Y, Baker T, et al. Network connectivity determines cortical thinning in early Parkinson’s disease progression. Nature Communications. 2018;9(12):1-10. doi:10.1038/s41467-017-02416-0 [3]: Marek K, Jennings D, Lasch S, et al. The Parkinson Progression Marker Initiative (PPMI). Progress in Neurobiology. 2011;95(4):629-635. doi:10.1016/j.pneurobio.2011.09.005 [4]: Aubert-Broche B, Fonov VS, Garc´ıa-Lorenzo D, et al. A new method for structural volume analysis of longitudinal brain MRI data and its application in studying the growth trajectories of anatomical brain structures in childhood. Neuroimage. 2013;82:393–402. doi: 10.1016/j. neuroimage.2013.05.065. [5]: La Joie R, Perrotin A, Barre L, et al. Region-Specific Hierarchy between Atrophy, Hypometabolism, and b-amyloid (Ab) load in Alzheimer’s disease Dementia. Journal of Neuroscience. 2012;32(46):16265-16273. doi:10.1523/JNEUROSCI.2170-12.2012 [6]: Cammoun L, Gigandet X, Meskaldji D, et al. Mapping the human connectome at multiple scales with diffusion spectrum MRI. Journal of Neuroscience Methods. 2012;203(2):386-397. doi:10.1016/j.jneumeth.2011.09.031 [7]: Mišić B, Betzel RF, Nematzadeh A, et al. Cooperative and Competitive Spreading Dynamics on the Human Connectome. Neuron. 2015;86(6):1518-1529. doi:10.1016/j.neuron.2015.05.035 [8]: Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:1-7. doi:10.1186/1471-2105-10-48 [9]: Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with PANTHER Classification System. Nat Protoc. 2013;8(8):1551-1566. doi:10.1038/nprot.2013.092
To cite this abstract in AMA style:
C. Tremblay, S. Rahayel, A. Vo, F. Morys, G. Shafiei, R. Markello, Z. Gan-Or, B. Misic, A. Dagher. Progression of brain atrophy in Parkinson’s disease is shaped by connectome and local vulnerability [abstract]. Mov Disord. 2021; 36 (suppl 1). https://www.mdsabstracts.org/abstract/progression-of-brain-atrophy-in-parkinsons-disease-is-shaped-by-connectome-and-local-vulnerability/. Accessed July 5, 2025.« Back to MDS Virtual Congress 2021
MDS Abstracts - https://www.mdsabstracts.org/abstract/progression-of-brain-atrophy-in-parkinsons-disease-is-shaped-by-connectome-and-local-vulnerability/