Objective: To build an AI model that can predict the monopolar review in deep brain stimulation (DBS) for Parkinson’s disease (PD) based on imaging.
Background: DBS is an effective therapy for PD. However, there remains inconsistency in outcomes across community populations. The brain location of the DBS lead, termed “lead location”, plays a major role in the overall “programmability” of the device and degree of clinical benefit obtained [1]. Programmability is determined via the “monopolar review” – a systematic survey of the DBS device.
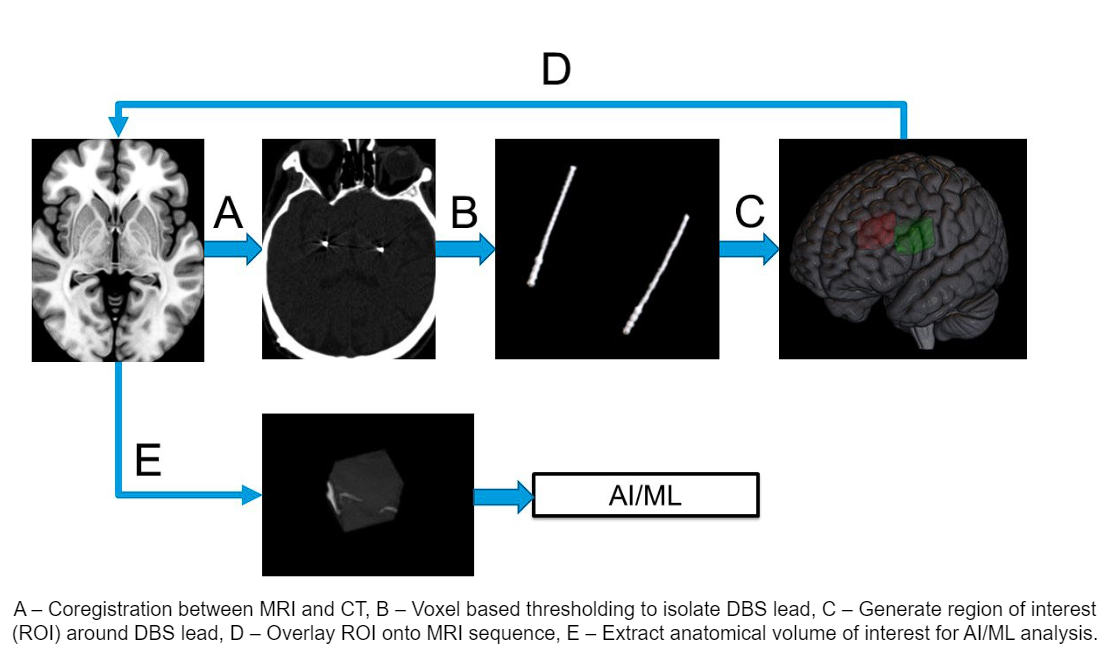
Method: Imaging data from 86 DBS implantation surgeries from 62 unique patients including preoperative MRI sequences were coregistered to the postoperative CT head using Advanced Normalization Tools. Followed by isolation of the DBS lead artifact from the CT head using voxelwise thresholding. Patient specific segmentation of the GPi was obtained for every patient using segmentation tool[2]. The different imaging modalities are combined to form the input for our AI model, illustrated in [figure 1]. We evaluated our model across a 5 fold cross-validation. Our performance metric was the average root mean square error (RMSE) of the thresholds across the four contacts in the DBS lead.
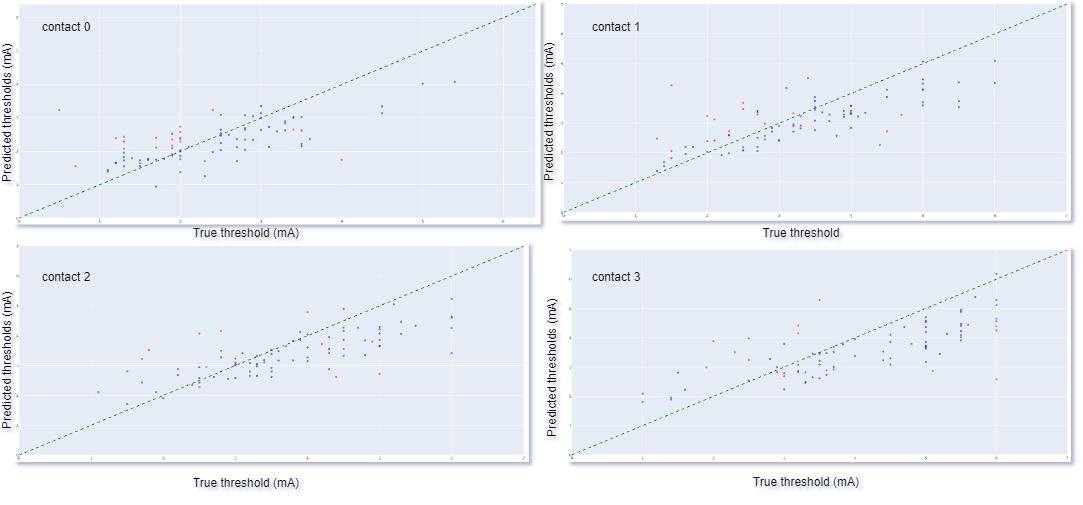
Results: We achieved a mean (SD) RMSE of 1.057 (0.2) mA, using Densenet 121 based 3D convolutional neural network (CNN) architecture. The true vs predicted thresholds of the 3D CNN for the training and the testing (validation) input of a fold in 4 contacts is plotted in [Figure 2]. The training threshold values indicate model fit to the training data while testing threshold values indicates model’s generalizability. Our 3D CNN model can predict monopolar review thresholds across 4 contacts of DBS electrode based on the trajectory of DBS electrode and relative anatomy in close proximity of the electrode.
Conclusion: Our results demonstrate the feasibility of medical imaging-based computer vision AI models as an alternative pathway to understanding the relationship between the DBS electric field and the volume of neuronal tissue activated. This method does not require resource intensive computational models and the associated assumptions made on the neural biophysical axon models to compute. This model sets the foundation for a pre-surgical decision-making tool that could significantly impact the surgical planning process for functional stereotactic neurosurgery.
Image processing workflow
CNN predictions, training: blue and testing: red.
References: [1] Holland, M. T., Jiao, J., Mantovani, A., Anderson, S., Mitchell, K. A., Safarpour, D., & Burchiel, K. J. (2022). Identifying the therapeutic zone in globus pallidus deep brain stimulation for Parkinson’s disease. Journal of Neurosurgery, 138(2), 329–336. https://doi.org/10.3171/2022.5.JNS22152 (Holland et al., 2022)
[2] Billot, B., Greve, D. N., Puonti, O., Thielscher, A., Van Leemput, K., Fischl, B., Dalca, A. V., & Iglesias, J. E. (2023). SynthSeg: Segmentation of brain MRI scans of any contrast and resolution without retraining. Medical Image Analysis, 86, 102789. https://doi.org/10.1016/j.media.2023.102789
To cite this abstract in AMA style:
V. Lavu, M. Godhala, T. Batchali, S. Aghili-Mehrizi, J. Wong. Prediction of The Monopolar Review in Deep Brain Stimulation for Parkinson’s Disease using Imaging [abstract]. Mov Disord. 2024; 39 (suppl 1). https://www.mdsabstracts.org/abstract/prediction-of-the-monopolar-review-in-deep-brain-stimulation-for-parkinsons-disease-using-imaging/. Accessed July 10, 2025.« Back to 2024 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/prediction-of-the-monopolar-review-in-deep-brain-stimulation-for-parkinsons-disease-using-imaging/