Category: Parkinsonism, Atypical: MSA
Objective: To automatically distinguish between healthy controls (HC) and patients with multiple system atrophy (MSA) with a completely automatic approach using a 3D convolutional neural network (CNN) and MRI maps as input.
Background: The differentiation of parkinsonism has benefited from machine learning approaches with great promise, including some deep learning applications [1], [2], [3], [4]. Despite the limited sample size due to the rarity of MSA, we proposed a 3D whole-brain analysis performed by a CNN based on two different MRI inputs.
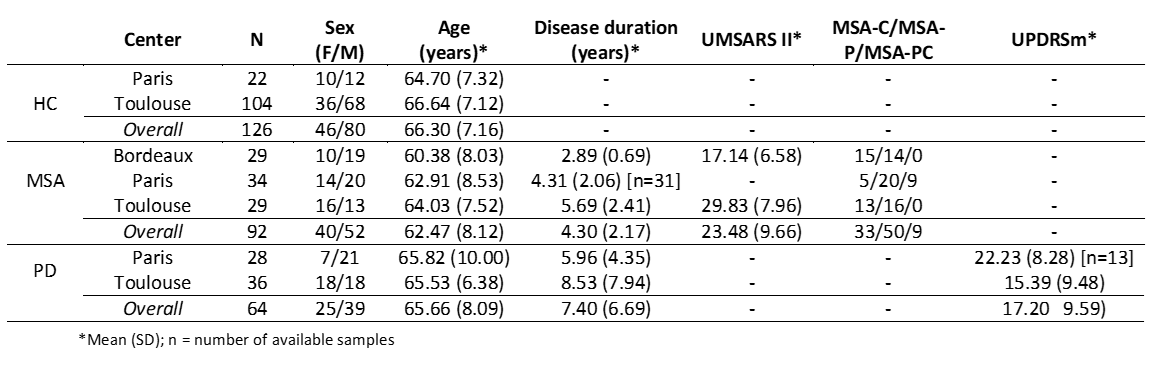
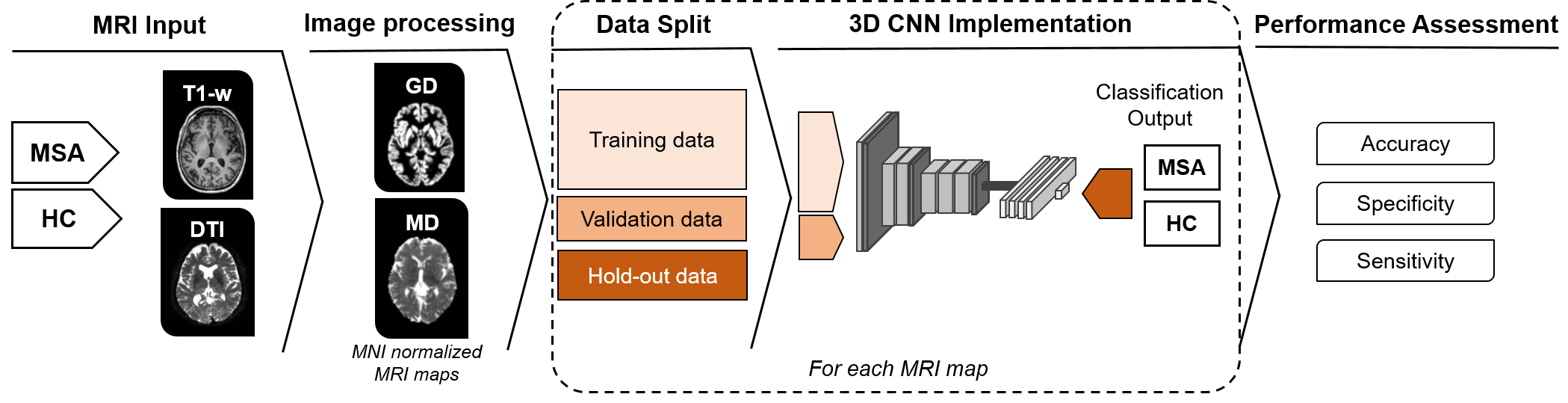
Method: We collected MRI data from three French reference centers for MSA, including 126 HC and 92 MSA patients (see [table1]). Image processing included: (i) computation of gray density probability (GD) maps from T1-weighted and mean diffusivity (MD) maps from diffusion tensor imaging [5]; (ii) spatial normalization to the MNI template. We trained a 3D CNN [6] with each type of whole-brain MRI map and both maps to classify MSA patients and HC. [figure1] illustrates an overview of the proposed approach. Moreover, we explored CNN performance after stratifying MSA patients into different degrees of brain alterations based on MRI features. Finally, we used the same MSA-HC-trained CNN to test the discrimination between MSA and PD patients (n=64).
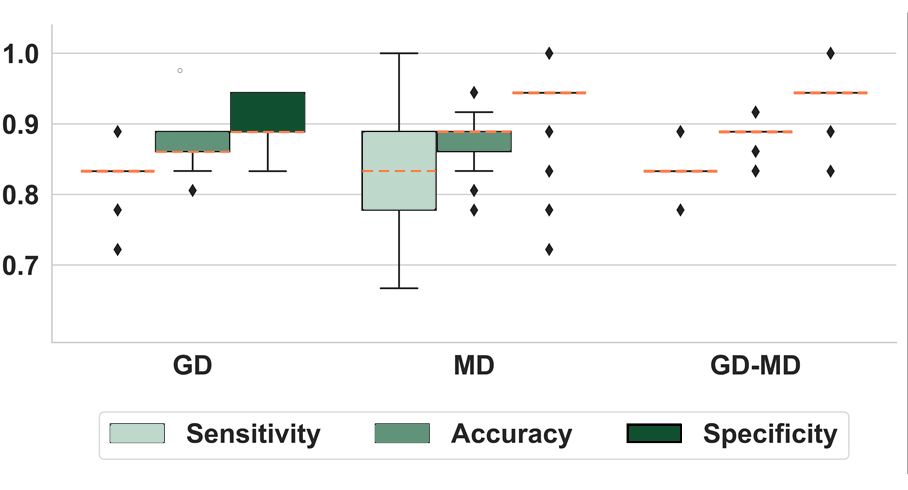
Results: MSA patients were distinguished from HC with high accuracy (> 0.85) using GD and MD separately (see [figure2]). MD achieved higher mean specificity (0.94 vs. 0.91 for GD) but more variable sensitivity compared to GD, although with a comparable mean value (0.83). The CNN trained with both GD and MD outperformed the single modality models. Training the model with a subset of MSA patients with milder image alterations allowed a better CNN generalizability compared to subsets with more severe alterations and not vice versa. Moreover, the MSA-HC-trained CNN classified PD patients with a high percentage (see [table2]).
Conclusion: Our findings support the feasibility of a deep learning approach for the automatic differentiation between HC and MSA patients using GD, MD, or both. Furthermore, we showed that an earlier disease stage may be more suitable to differentiate a later stage. In conclusion, the MSA-HC-trained CNN showed encouraging results for the differential diagnosis of Parkinsonian syndromes.
Demographics and clinical data of patients and HC
Methods Overview
Boxplots showing CNN performance (MSA vs. HC)
CNN Performance – Test on PD Patients as mean (SD)
References: [1] H. Zhao et al., “Deep learning based diagnosis of Parkinson’s Disease using diffusion magnetic resonance imaging,” Brain Imaging Behav, vol. 16, no. 4, pp. 1749–1760, Aug. 2022, doi: 10.1007/s11682-022-00631-y.
[2] S. Kiryu et al., “Deep learning to differentiate parkinsonian disorders separately using single midsagittal MR imaging: a proof of concept study,” Eur Radiol, vol. 29, no. 12, pp. 6891–6899, Dec. 2019, doi: 10.1007/s00330-019-06327-0.
[3] M. Camacho et al., “Explainable classification of Parkinson’s disease using deep learning trained on a large multi-center database of T1-weighted MRI datasets,” Neuroimage Clin, vol. 38, Jan. 2023, doi: 10.1016/j.nicl.2023.103405.
[4] L. Chougar et al., “Automated Categorization of Parkinsonian Syndromes Using Magnetic Resonance Imaging in a Clinical Setting,” Movement Disorders, vol. 36, no. 2, pp. 460–470, Feb. 2021, doi: 10.1002/mds.28348.
[5] P. Péran et al, “MRI supervised and unsupervised classification of Parkinson’s disease and multiple system atrophy,” Movement Disorders, vol. 33, no. 4, pp. 600–608, Oct. 2018, doi: 10.1002/mds.27307.
[6] G. M. Mattia et al., “Multimodal MRI-Based Whole-Brain Assessment in Patients In Anoxoischemic Coma by Using 3D Convolutional Neural Networks,” Neurocrit Care, vol. 37, no. S2, pp. 303–312, Aug. 2022, doi: 10.1007/s12028-022-01525-z.
To cite this abstract in AMA style:
GM. Mattia, L. Chougar, WG. Meissner, A. Foubert-Samier, A. Pavy-Le Traon, O. Rascol, M. Fabbri, S. Lehéricy, P. Péran. A 3D CNN Approach for the Classification of Multiple System Atrophy using Multimodal Multicentric MRI [abstract]. Mov Disord. 2024; 39 (suppl 1). https://www.mdsabstracts.org/abstract/a-3d-cnn-approach-for-the-classification-of-multiple-system-atrophy-using-multimodal-multicentric-mri/. Accessed June 30, 2025.« Back to 2024 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/a-3d-cnn-approach-for-the-classification-of-multiple-system-atrophy-using-multimodal-multicentric-mri/